Floyd-Warshall
| Tags | CS161Graphs |
|---|
The motivation
What if we wanted to find all pairwise shortest path? (APSP)?
We can use Bellman-Ford steps, which takes complexity. If there are no negative edges, we can use steps of djikstra’s algorithm, which takes .
If , then BF takes complexity, and Djikstra takes , which is OK but can we just do it in steps, regardless of edge weight sign?
Implementation
Substructure
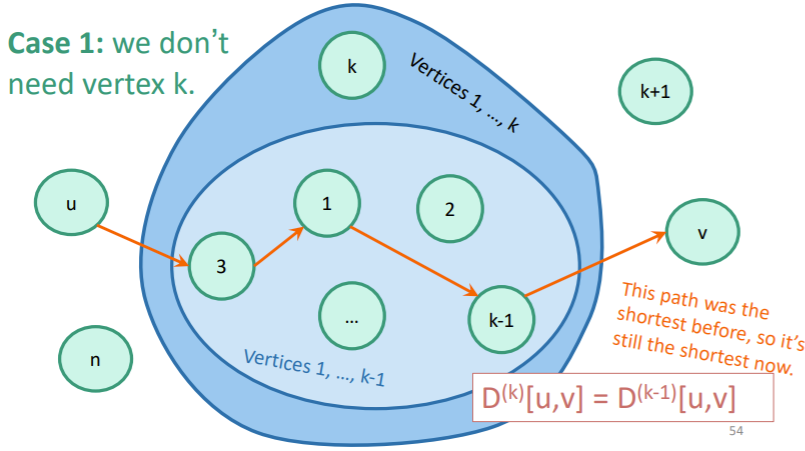
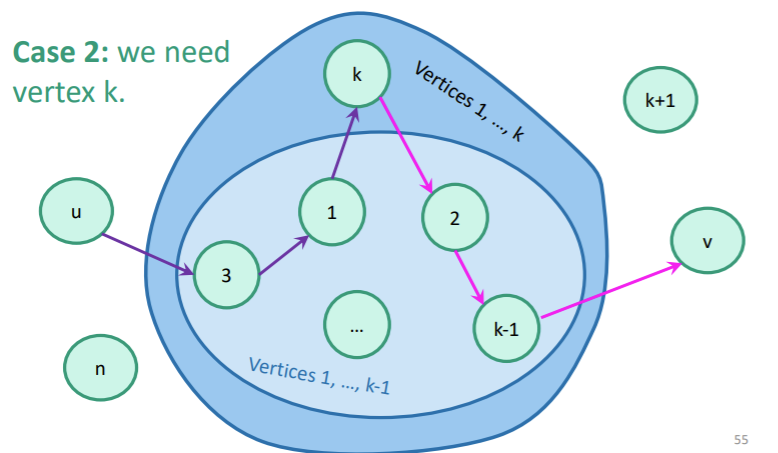
Here, we put on our dynamic programming thinking caps. What sort of subproblem can we design? How about an array that is the shortest path between such that all the vertices between are vertices .
This actually a little bit counterintuitive. The knee-jerk adaptation is to use bellman-ford but pairwise. In that case, would be the shortest path such that the number of vertices between are less than . However, you can show to yourself that this yields complexity and is not good.
What happens at each step
You would initialize as the weighted adjacency matrix (and non-edges are infinite). This because direct edges have no vertices between them, which satisfies the definition of . Then, you would do the following:
- Select a pair of nodes (represented as an element in ).
- Try if is of lower weight than . If you can find one, update the distance in . If not, keep what you have. You can use and as defined in your .
Inductively, it should be easy to show that you keep the inductive definition of after doing this step.
Much like Bellman-Ford, you can show that you only need steps to complete. In each step, you iterate through pairs of vertices, yielding a final complexity of .
Algorithm box

Detecting negative cycles
If there are negative cycles, then . Because negative cycles necessairly mean that you can circle back to where you started and be better off.